George Murray
Predoctoral Associate
I work with mouse models of peripheral neuropathy to identify disease mechanisms and gene expression signatures to provide insights that may lead to treatment strategies.
George C. Murray is a PhD candidate studying diseases of the peripheral nervous system in the lab of Dr. Robert Burgess at the Jackson Laboratory in Bar Harbor, ME. His bioinformatics-oriented research involves comparative and integrative analyses of data across multiple ‘omics modalities such as RNA sequencing and metabolomics to uncover predictive signatures of disease, characterize mouse models of disease, and identify therapeutic targets. George’s research also has a strong wet-lab component, which involves validation of a potential therapeutic target using mouse models, and mapping disease-modifying genes in genetically diverse populations of mice. Aside from research, George actively mentors undergraduate research fellows at The Jackson Laboratory and has served in several leadership roles throughout graduate school.
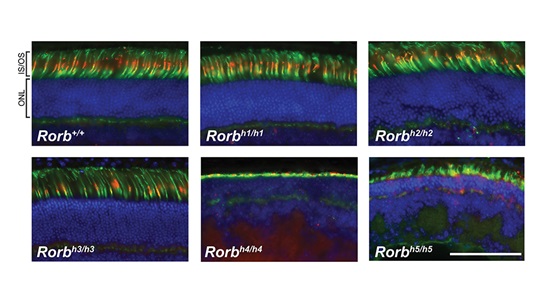
George is involved in several projects: 1) Quantitative trait loci mapping of genes that modify Charcot-Marie-Tooth (CMT) type 2D phenotypes in validated CMT mouse models and diversity outbred mice. 2) Assessment of Sipa1l2 in the C3-PMP22 mouse model as a candidate modifier and therapeutic target for CMT1A identified by patient-only GWAS. 3) Comparative analysis of gene expression datasets from numerous CMT models to develop a phylotranscriptomic tree of CMT models and identify prognostic gene expression signatures and therapeutic strategies. 4) Characterization of an allelic series of spontaneous Rorb mutant mice with a gait phenotype and retinal abnormalities. 5) Metabolomic- and gene expression-based characterization of a mouse model of NADK2 deficiency. Together, these projects illustrate the utility of mice for therapeutic target identification, disease modeling, and validation studies – the major narrative theme of George’s dissertation.