A team of JAX researchers recently published “The Genome of C57BL/6J "Eve", the Mother of the Laboratory Mouse Genome Reference Strain”, the female founder of the C57BL/6J (B6J) currently distributed by The Jackson Laboratory. The B6J strain is maintained via a Genetic Stability Program with periodic reintroduction of cryorecovered mice derived from a single breeder pair. The genomic sequence of Eve more accurately represents the genome of today’s C57BL/6J mice compared to the first release of the mouse reference genome (GRCm38) 20 years ago.
The point of using mice for biomedical research
The mouse is an essential model organism for biomedical research. Decades of research analyzing and manipulating the mouse genome have translated into a better understanding of human physiology and diseases. Genomic approaches have the ability to connect phenotypes with genotypes for a wide variety of traits and to use the resulting molecular insights to develop new ways to cure and prevent disease. The laboratory mouse occupies a central place in this vision as a well-characterized organism for modelling human disease states.
The availability of the full mouse genome sequence greatly advanced both the type and the pace of discovery. Mus musculus, a species of mouse, has been one of the five key model organisms sequenced since the beginnings of the Human Genome Project. The sequence of the mouse genome is a key informational tool for understanding the contents of the human genome and a key experimental tool for biomedical research. The initial sequence of the mouse genome reported in 2002 was a first step in this intellectual endeavor (Waterston et al., 2002) and allowed comparative analysis to examine the similarities and differences between the human and mouse genomes.
Isogenic laboratory mouse strains enhance reproducibility because individual animals are genetically identical. For the most widely used isogenic strain, C57BL/6, there exists a wealth of genetic, phenotypic, and genomic data, including a high-quality reference genome that evolved with time (GRCm38.p6 being the latest release). However, traditional inbreeding practices create a genetic bottleneck: every generation, between 10 and 30 new mutations pop up and are passed on to offspring. Unfortunately, most of these mutations are not easy to spot until researchers are investigating specific traits (Reardon, 2017). It is now very clear that different suppliers of B6 mice have inadvertently created divergent substrains when they obtained descendants of C57BL/6J mice and bred them in seclusion. This limitation, driven by genetic drift, often contributes to unexpected phenotypes and hampers the reproducibility of data generated with laboratory mice. Although most mutations go unnoticed, some occur in genes that affect a mouse's appearance or physiology (Reardon, 2017). Therefore, the genomic sequences of B6 substrains are different from the corresponding Reference Genome Sequences.
What is a Reference Genome?
In very simple terms, a Reference Genome Sequence for any given organism is the complete and ordered “assembly” of DNA, as denoted by the nucleotides A, T, C, and G. Reference genome sequences result from the de novo sequencing and assembly of a haploid complement of an organism’s genome. A reference genome is the initial sequence to which all subsequent sequences are compared but it is often incomplete mainly due to technical constraints. The human and mouse gene catalogues remain imperfect. Both genome sequences are still incomplete. Some authentic genes are missing, fragmented or otherwise incorrectly described, and some predicted genes may be in fact pseudogenes. The first mouse reference genome, fully sequenced in 2002, contained approximately 24,500 protein-coding genes distributed across 19 pairs of autosomes, plus the X and Y sex chromosomes. Research groups around the world rely on reference sequences for much of current genetics research, as well as for designing and validating new mouse models built using specific genetic backgrounds.
The goal of reference genomes is to provide the most accurate and complete representation of specific genomes by correcting errors, closing gaps, finding ways to represent difficult regions such as centromeres, and capturing genetic variation (McGarvey et al., 2015). Since the completion of the Mouse Genome Project in 2002, the mouse reference genome has continued to be updated and refined by the Genome Reference Consortium (GRC), a team of scientists from several institutions in Europe and in USA. The GRC periodically re-annotate the mouse genome as the C57BL/6J assembly is updated, the annotation pipeline software is further improved on, or as additional data become available.
The Genome Reference Consortium Mouse Build 38 patch release 6 (GRCm38.p6) is the latest version released in September, 2017 representing the C57BL/6J strain. Despite being one of the highest quality mammalian genome assemblies ever produced, it still has more than 600 gaps and includes sub-optimal representations for some genes. GRCm38 still contains 523 gaps within chromosome sequences, and there are nearly 300 unresolved issues that have been reported to the GRC (Sarsani et al., 2019). In addition to gaps, these issues include reports of localized sequence mis-assembly, missing genic and non-genic sequences, sequencing errors and suspect variation. These types of assembly issues inflate false positive rates in reference-based variant calling.
In general, the preference is to use the latest version of a given species reference genome, except where bioinformatics processing tools or resources are not compatible, or to keep consistency in a long-term project and for comparison purposes.
“Eve” has been sequenced
JAX researchers recently provided an update to the mouse reference genome that more accurately represents the genome of today’s C57BL/6J mice, using long read, short read, and optical mapping technologies to generate a de novo assembly of the C57BL/6J Eve genome. They have identified structural variations, closed gaps in the mouse reference assembly, and revealed previously unannotated coding sequences (Sarsani et al., 2019). To generate data for their de novo assembly of the C57BL/6J Eve genome, they used a range of technologies, including; Pacific BioSciences long read technology at 66X whole genome coverage, Illumina short read at 32X whole genome coverage, and Bionano Genomics optical maps.
Eve's genome is very different from the 2002 mouse reference genome. The authors found over 500, high quality, recurring variants (SNP/Indels) and over 40 structural variants (inversions, deletions, and duplications) involving protein-coding genes compared to the prior reference genome GRCm38.p6 (Figure 1).
The new sequencing data from Eve C57BL/6J (a much more realistic representation of current B6J mice from The Jackson Laboratory) will be incorporated in the upcoming release of the Mouse Genome Reference sequence “GRCm39” planned for the end of 2019/early 2020. This will be an invaluable tool for researchers and mutant/transgenic cores as they design and validate new models in the C57BL/6J genetic background.
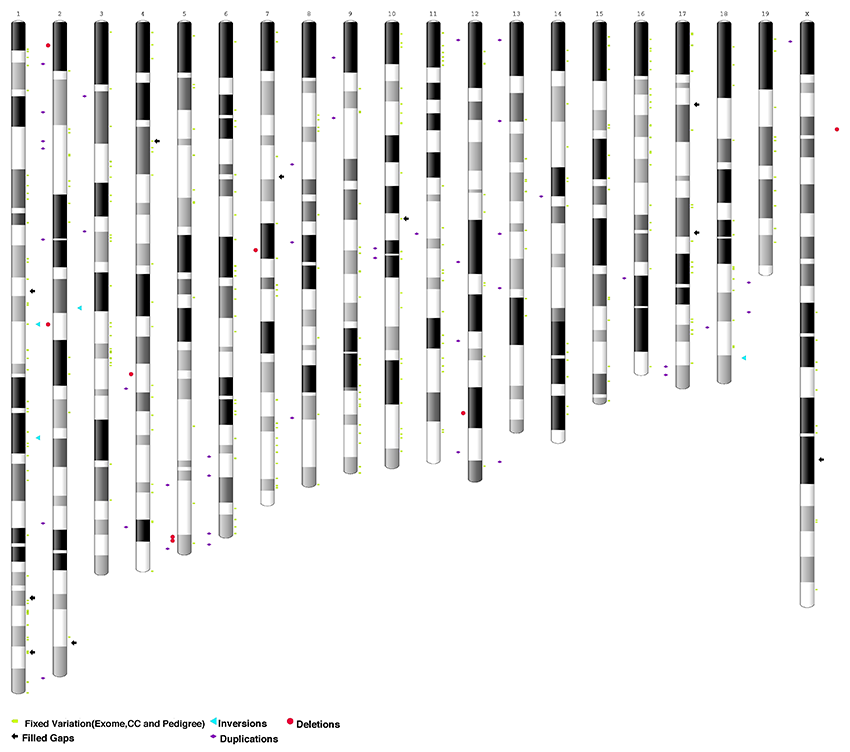
Figure 1. Ideogram of GRCm38 assembly annotated to highlight resolved gaps (vs. current reference), structural variants, and fixed variation using B6J Eve data. Adapted from Sarsani et al 2019. |
Future generations of C57BL/6J mice will remain much closer to Genome Reference sequence GRCm39.
B6J mice are living reagents subject to genetic drift (as are all inbred strains), an unavoidable source of accumulating genetic variability that can have an impact on reproducibility over time. Nearly 20 years after the first release of the mouse reference genome, individuals from the strain it represents (C57BL/6J) are at least 26 inbreeding generations removed from the breeders used to generate the mouse reference genome GRCm38 (Figure 2). Under the highly selective breeding paradigms employed for inbred laboratory strains, this genetic distance (>20 generations) is sufficient for rapid fixation of 98.7% of variants, such that today’s C57BL/6J mice are by definition a sub-strain of the animals from which GRCm38 is derived.
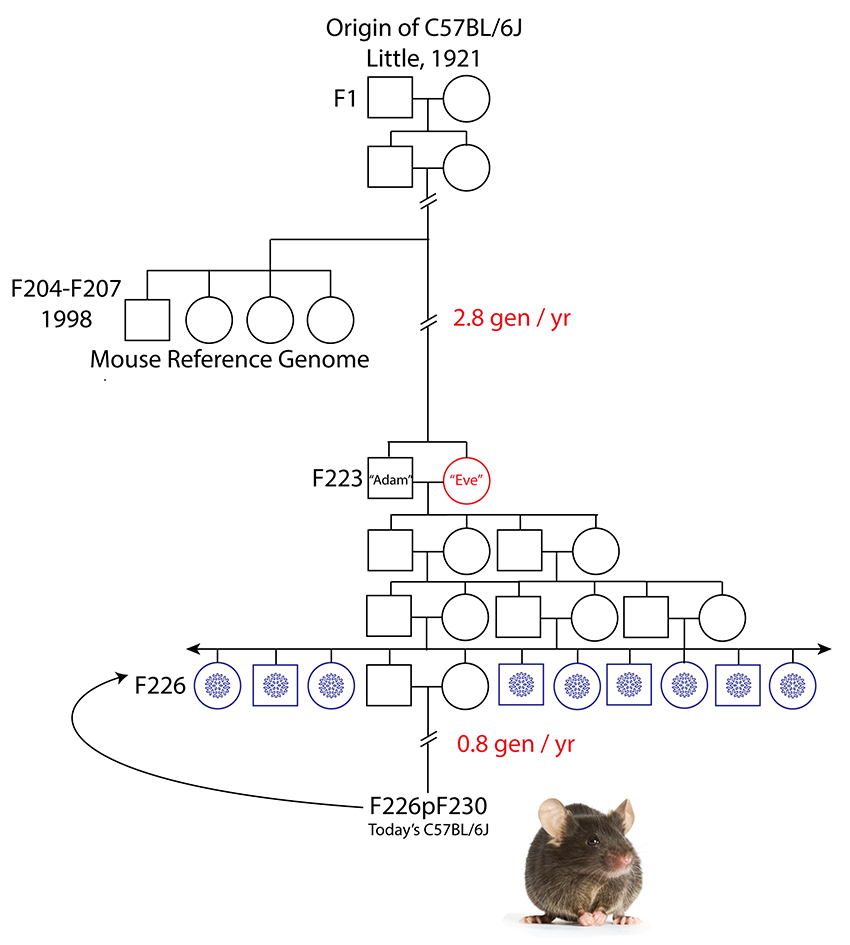
Figure 2. Origin of the inbred strain C57BL/6J. Inbred laboratory mouse strains are maintained by brother x sister mating. Filial (F) generations from which mice contributing to the reference assembly clone libraries and from which the B6Eve mouse were derived are shown. Cryopreserved embryo stock is represented by blue snowflakes at F226, 3 generations from Adam and Eve at F223. Generations subsequent to the cryopreservation event are F226p###, e.g., F226p230, which means embryos cryopreserved at F226 were recovered and there were an additional 4 generations of subsequent inbreeding. Adapted from Sarsani et al 2019. |
However, since 2003, The Jackson Laboratory manages the rate of genetic drift by selling only B6J mice descended from the breeder pair so-called “Adam and Eve” by Sarsani and collaborators (Sarsani et al., 2019). This was accomplished by freezing thousands of embryos of their grandchildren (Figure 2), enough to last for 25-30 years (Reardon, 2017), and which are periodically used to replenish pedigreed foundation breeding colonies (Wiles and Taft, 2010). The Jackson Laboratory’s unique, patented Genetic Stability Program (GSP) effectively limits cumulative genetic drift, including that caused by copy number variation (CNV), by rebuilding our foundation stocks from cryopreserved, pedigreed embryos every five generations. This process introduces a controlled bottleneck that minimizes the accumulation of genetic change. Therefore, B6J mice obtained from JAX will be a maximum of 7 generations removed from Eve (Reference sequence GRCm39).
These findings point to the genetic distance between the original B6J mice sequenced in 2002 compared to Eve’s within JAX. In addition, they illustrate the risks of substrain divergence when researchers or vendors maintain colonies using “traditional” inbreeding methods without programs to minimize the accumulation of spontaneous mutations through generations whose sequence will not correspond tightly to the Reference Genome data.
Learn more about additional advantages of using JAX mice at the Why JAX Mice page.
Recommended Resources
Webinar: Protect Your Research Know Your B6 Mouse
Webinar: Achieving Reproducible Mouse Studies
Remember only YOU can prevent genetic drift
References
Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002 Dec 5;420(6915):520-62. [PMID: 12646614]
Genome Reference Consortium Mouse Build 38 patch release 6 (GRCm38.p6)
McGarvey KM, Goldfarb T, Cox E, et al. Mouse genome annotation by the RefSeq project. Mamm Genome. 2015;26(9-10):379–390. doi:10.1007/s00335-015-9585-8. [PMID: 26215545]
Reardon S. 2017. Lab mice's ancestral ‘Eve’ gets her genome sequenced. Nature. 2017 Nov 13;551(7680):281. doi: 10.1038/nature.2017.22974. [PMID: 29144484]
Sarsani et al 2019. The Genome of C57BL/6J "Eve", the Mother of the Laboratory Mouse Genome Reference Strain. G3 (Bethesda). 2019 Jun 5;9(6):1795-1805. doi: 10.1534/g3.119.400071. [PMID: 30996023]
- Genomic version LXEJ02000000: Mus musculus strain C57BL/6J isolate Eve, whole genome shotgun sequencing project consists of sequences LXEJ02000001-LXEJ02012661 GenBank: LXEJ00000000.2 https://www.ncbi.nlm.nih.gov/nuccore/LXEJ02000000
- B6Eve assembly along with annotation and an assembly hub: ftp://ftp.jax.org/b6eve
- Visualization of the assembly: https://genome.ucsc.edu → MyData → Track Hubs → My Hubs with the following URL: ftp://ftp.jax.org/b6eve/assemblyhub/hub.txt
Wiles and Taft 2010. The sophisticated mouse: protecting a precious reagent. Methods Mol Biol. 2010;602:23-36. doi: 10.1007/978-1-60761-058-8_2. [PMID: 20012390]